Linear Models (lm)
Table of Contents
- Overview
- Fit a model with no intercept
- Interpret coefficients of categorical variables in
lm confint(object, parm, level = 0.95, …)- Calculate confidence intervals for model parameters
- Draw Fitted versus Residuals Plot
- Draw Normal QQ Plot
lmtest::bptest(model, ...)- Perform BP test
shapiro.test(x)- Perform Shapiro-Wilk Test
hatvalues(model, ...)- Calculate Leverage
rstandard(model, ...)- Calculate Standardized Residual
cooks.distance(model, ...)- Find influential observations
- Calculate Variance Inflation Factors
- Calculate Partial Correlation Coefficient
- Perform a model selection with exhaustive search
step(object, scope, direction)- Perform a model selection with stepwise search based on AIC/BIC
- Calculate AIC, BIC
- Perform Leave-One-Out Cross Validation
- Extract the number of observations from a fit
- Change the null hypothesis to \(H_0 = T\) in
summary(lm)
Overview
anova(object_1, object_2)- Compare a submodel with an outer model and produce an analysis of variance table.
coef(object)- Extract the regression coefficient (matrix). Long form:
coefficients(object). deviance(object)- Residual sum of squares, weighted if appropriate.
formula(object)- Extract the model formula.
plot(object)- Produce four plots, showing residuals, fitted values and some diagnostics.
- predict(object, newdata=data.frame)
- The data frame supplied must have variables specified with the same labels as the original. The value is a vector or matrix of predicted values corresponding to the determining variable values in
data.frame. print(object)- Print a concise version of the object. Most often used implicitly.
residuals(object)- Extract the (matrix of) residuals, weighted as appropriate. Short form:
resid(object). step(object)- Select a suitable model by adding or dropping terms and preserving hierarchies. The model with the smallest value of AIC (Akaike’s An Information Criterion) discovered in the stepwise search is returned.
summary(object)- Print a comprehensive summary of the results of the regression analysis.
vcov(object)- Returns the variance-covariance matrix of the main parameters of a fitted model object.
Fit a model with no intercept howto
You can use this feature for Parameterization, which makes the resulting coefficients independent.
(Intercept) Bwt SexM
-0.41495263 4.07576892 -0.08209684
Bwt SexF SexM
4.0757689 -0.4149526 -0.4970495
Interpret coefficients of categorical variables in lm howto
Consider the following model: \[ Y = \beta_0 + \beta_1 x_1 + \beta_2 x_2 + \beta_3 x_1 x_2 + \epsilon \]
(Intercept) hp am hp:am
26.6248478696 -0.0591369818 5.2176533777 0.0004028907
In this case, am is a categorical variable, which determine the observation is automatic or manual.
(Intercept)- is the estimated average mpg for a car with an automatic transmission and 0 hp.
(Intercept) + am- is the estimated average mpg for a car with a manual transmission and 0 hp.
am- is the estimated difference in average mpg for cars with manual transmissions as compared to those with automatic transmission, for any hp.
hp- is the estimated change in average mpg for an increase in one hp, for either transmission types.
hp:am- is the estimated difference in the change in average mpg for an increase in disp, for a car of a certain hp (or vice versa).
confint(object, parm, level = 0.95, …) reference
Calculate confidence intervals for model parameters howto
2.5 % 97.5 %
(Intercept) -0.774822875 2.256118188
disp -0.002867999 0.008273849
hp -0.001400580 0.011949674
wt 0.380088737 1.622517536
am -0.614677730 0.926307310
2.5 % 97.5 %
wt 0.3800887 1.622518
Draw Fitted versus Residuals Plot howto plot
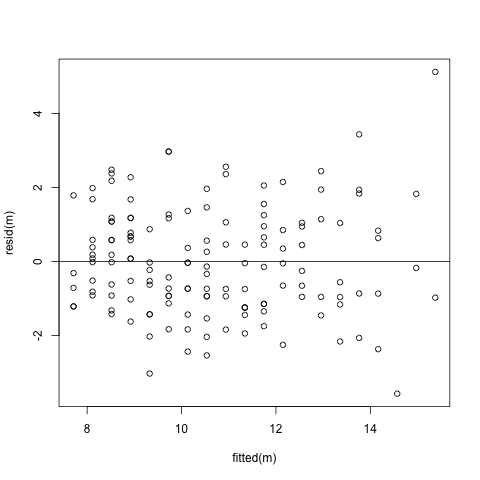
- Linearity
- Residuals should be centered at 0.
- Equal Variance
- Variances are similar across fitted values.
Draw Normal QQ Plot howto plot
- QQ stands for Quantile-Quantile.
- X axis for theoretical quantiles, Y axis for sample quantiles.
As for checking normality of errors:
- X axis goes to the normal distribution quantiles
- Y axis goes to the residuals of the model.
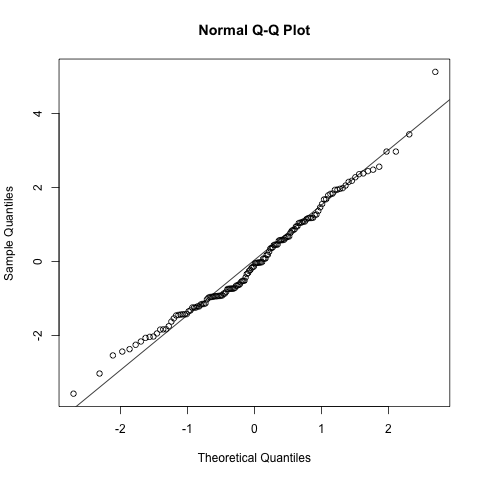
qqplot = function(e) {
n = length(e)
# For n = 10, this vector contains quantiles of [0.05 0.15 0.25 ... 0.95]
normal_quantiles = qnorm(((1:n - 0.5) / n))
# plot theoretical verus observed quantiles
plot(normal_quantiles, sort(e))
# calculate line through the 1st and 3rd quartiles
slope = (quantile(e, 0.75) - quantile(e, 0.25)) / (qnorm(0.75) - qnorm(0.25))
intercept = quantile(e, 0.25) - slope * qnorm(0.25)
# add to existing plot
abline(intercept, slope, lty = 2, lwd = 2, col = "dodgerblue")
}lmtest::bptest(model, ...) reference
Perform BP test howto
- Test homoscedasticity. (whether or not the error is equally distributed)
studentized Breusch-Pagan test
data: m
BP = 9.5294, df = 1, p-value = 0.002022
shapiro.test(x) reference
Perform Shapiro-Wilk Test howto
- Unlike other test functions,
shapiro.testaccepts avectorinstead of almobject.
Shapiro-Wilk normality test
data: rnorm(25)
W = 0.91987, p-value = 0.0509
hatvalues(model, ...) reference
Calculate Leverage howto
[1] 0.04048627 0.04048627 0.05102911 0.04048627 0.03675069 0.04317538
[7] 0.09757509 0.08046517 0.04958291 0.03510034 0.03510034 0.03886509
[13] 0.03886509 0.03886509 0.05458370 0.06327290 0.07888001 0.07592585
[19] 0.09277415 0.07704010 0.04819161 0.03132530 0.03132530 0.09757509
[25] 0.03675069 0.07592585 0.05253020 0.03903750 0.12568847 0.03675069
[31] 0.27459288 0.04099664
rstandard(model, ...) reference
Calculate Standardized Residual howto
Calculates: \[ r_i = \frac{e_i}{s_e\sqrt{1 - h_i}} \]
[1] -0.42118619 -0.42118619 -0.25341558 -0.31547671 0.14271781 -1.27952886
[7] 0.24990523 -0.39648860 -0.21698865 -0.66062513 -1.02957426 -0.37436332
[13] -0.13671767 -0.69122419 -1.52076121 -1.34530901 0.07920301 1.83232949
[19] 1.04609436 2.21923491 -0.52556608 -1.14798677 -1.22689309 -0.02259947
[25] 0.27459819 0.45893182 0.56112724 2.11548290 1.02806065 0.40647857
[31] 2.35785298 -0.33359639
cooks.distance(model, ...) reference
Find influential observations
Calculates and test the value heuristically:
\[\begin{aligned} D_i &= \frac{1}{p}r_i^2\frac{h_i}{1 - h_i}\\ D_i &> \frac{4}{n} \quad \text{(when it's true, the observation is influential)} \end{aligned} \] [1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[13] FALSE FALSE FALSE FALSE FALSE TRUE FALSE TRUE FALSE FALSE FALSE FALSE
[25] FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE
Calculate Variance Inflation Factors howto
You can use car::vif(model). It appears that many packages include this kind of functions.
disp hp
2.670938 2.670938
Also, we can calculate it manually:
m = lm(mpg ~ disp + hp, data = mtcars)
md = lm(disp ~ hp, data = mtcars)
mh = lm(hp ~ disp, data = mtcars)
c(1 / (1 - summary(md)$r.squared), 1 / (1 - summary(mh)$r.squared))[1] 2.670938 2.670938
If the VIF of a parameter is larger than 5, heuristically it can be considered to be somewhat collinear to other variables.
Calculate Partial Correlation Coefficient howto
[1] -0.3258012
When this value is large, we can think that the unexplained portion of the candidate predictor, hp in the example above, and that of the response are highly correlated. This means that there is no significant collinearity between the candidate predictor and the existing predictors, disp in this case. So, adding the candidate predictor is of benefit.
We can also visualize this relationship:
plot(resid(my) ~ resid(mh))
abline(h = 0, lty = 2)
abline(v = 0, lty = 2)
abline(lm(resid(my) ~ resid(mh)))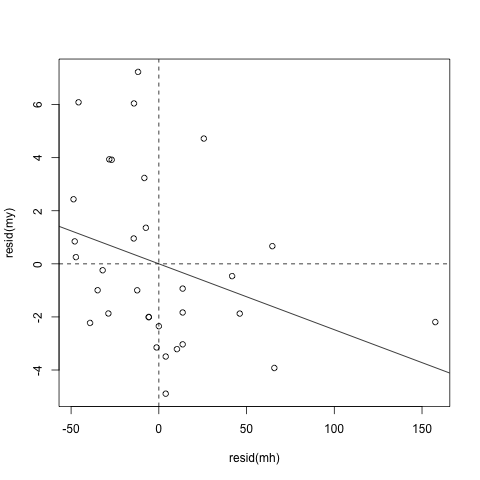
Perform a model selection with exhaustive search howto
- Use
leaps::regsubsets(formula, data = data) - This returns a list of best models of each size.
step(object, scope, direction) reference
Perform a model selection with stepwise search based on AIC/BIC howto
# Backward AIC
selected_model = step(full_model, direction = "backward")
# Backward BIC
selected_model = step(full_model, direction = "backward", k = log(n))
# Forward AIC
selected_model = step(start_model, scope = formula, direction = "forward")
# Both AIC
selected_model = step(start_model, scope = formula, direction = "both")Note that it is not available to use . in scope. To use . for the scope, you can pass a list to scope as follows:
step function respects the model hierarchy. For example, when we have two-interaction, both first order term should be in the model. Or, when we have a quadratic term, the first order term should be in the model.
Note that step() implicitly refers the dataset which the start_model bases on. So, if you use step() in a function, you should do it as follows:
step_backward = function(start_model) {
# NOTE: `step()` function implicitly refers the model's data by the name.
# Generally the data is accessible from the calling frame.
# So deines a new environment from it.
e = new.env(parent = parent.frame())
# Pass the arguments to the new `e`
e$start_model = start_model
with(e, {
n = nobs(start_model)
list(
aic = step(start_model, direction = "backward", trace = 0),
bic = step(start_model, direction = "backward", trace = 0, k=log(n))
)
})
}Calculate AIC, BIC howto
[1] 4.0000 274.2418
[1] 4.0000 280.7922
It returns c(<equivalent degrees of freedom>, <AIC>).
Perform Leave-One-Out Cross Validation howto
Extract the number of observations from a fit howto
Change the null hypothesis to \(H_0 = T\) in summary(lm) howto
By default, summary() calculates p-value of \(H_0: \beta = 0\). So we need to modify the formula for the model as follows: